|
 SVGM-exp β-v.1.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, 2021.
SVGM-exp β-v.1.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, 2021.
 Viruses β-v.4.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, v.1 - March 2020.
Viruses β-v.4.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, v.1 - March 2020.
 E-mail: rachedi@bioinformaticstools.org E-mail: rachedi@bioinformaticstools.org
|
|

|
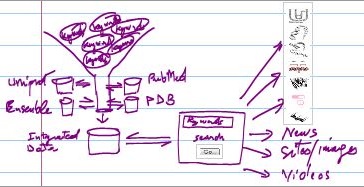
Disclaimer: The Viruses Database system offers data as they are from sources(*) used in the data integration. The data are by no mean complete about all types viruses, however, the database is regularly updatad and
the tool system is continuously further developed.
(*)Primarily the COVID-19 Genomics UK Consortium (COG-UK), NCBI/Genbank, EMBL genomics data & PDB structural data. |
References:
- Mutations in the SARS-CoV-2 complete genome sequences from strains isolated in Blida province, Algeria.
Abdelkrim Rachedi, JNBGP 2020: Journees Nationales
virtuelles de bioinformatique: Genomique et proteomique
- USDB, October 09-10, 2020 - Refer to
DOI: 10.13140/RG.2.2.20838.45123
 The Keynote Conference
presentation (1h:30Min) is available here: https://www.youtube.com/watch?v=JaIU6ufeI4g&t=1690s The Keynote Conference
presentation (1h:30Min) is available here: https://www.youtube.com/watch?v=JaIU6ufeI4g&t=1690s
- The Viruses, data annotation & integration tool, created by the DIRC Data Integration Module, is based on an improved version of the algorithm reported in the following research
paper:
Abdelkrim Rachedi et al., GABAagent: a system for integrating data on GABA
receptors. Bioinformatics. 2000 Apr;16(4):301-12.
|
|


 Viruses β-v.4.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, v.1 - March 2020.
Viruses β-v.4.0 -July 2021, © Abdelkrim Rachedi at the Dept. of Biology, Saida University, Algeria, v.1 - March 2020.
 The Keynote Conference
presentation (1h:30Min) is available here:
The Keynote Conference
presentation (1h:30Min) is available here: